Mutation Matrix Report¶
Here, we show describe the procedure generate Mutation Matrix report using sample data [*].
| [*] | The sample data is equipped with the example directory of the paplot directory. |
1. Minimal dataset¶
For generating Mutation Matrix Report using paplot, at least sample ID (Sample), gene name (Gene) and mutation type (MutationType) are required.
Sample,MutationType,Gene
SAMPLE00,intronic,GATA3
SAMPLE00,UTR3,CDH1
SAMPLE00,exonic,GATA3
SAMPLE01,splicing,WASF3
SAMPLE01,intronic,WASF3
SAMPLE01,exonic,NRAS
SAMPLE02,intronic,FBXW7
SAMPLE02,intronic,GATA3
SAMPLE02,ncRNA_intronic,ACVR2B
SAMPLE03,exonic,CAP2
SAMPLE03,intronic,PIK3CA
SAMPLE03,downstream,SEPT12
Although the column names are Sample, MutationType, and Gene, they can be arbitrary changed.
Set the column names in the [result_format_mutation] section of the configuration file.
[result_format_mutation]
col_group = MutationType
col_gene = Gene
col_opt_id = Sample
Then, execute the paplot.
paplot mutation {unzip_path}/example/mutation_minimal/data.csv ./tmp mutation_minimal \
--config_file {unzip_path}/example/mutation_minimal/paplot.cfg
2. Without header¶
SAMPLE00,intronic,GATA3
SAMPLE00,UTR3,CDH1
SAMPLE00,exonic,GATA3
SAMPLE01,splicing,WASF3
SAMPLE01,intronic,WASF3
SAMPLE01,exonic,NRAS
SAMPLE02,intronic,FBXW7
SAMPLE02,intronic,GATA3
SAMPLE02,ncRNA_intronic,ACVR2B
SAMPLE03,exonic,CAP2
SAMPLE03,intronic,PIK3CA
SAMPLE03,downstream,SEPT12
When the input data has no header (column names), it is necessary to set the column number to each key in the [result_format_mutation] section of the configuration file.
[result_format_mutation]
# Set the value of the header option to False
header = False
col_group = 2
col_gene = 3
col_opt_id = 1
Then execute paplot.
paplot mutation {unzip_path}/example/mutation_noheader/data.csv ./tmp mutation_noheader \
--config_file {unzip_path}/example/mutation_noheader/paplot.cfg
3. Customizing pop-up information¶
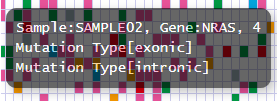
We can customize the pop-up information that appears upon mouseover events. In the minimal dataset, the pop-up information displays sample, gene, and mutation type as illustrated below:
Before customization
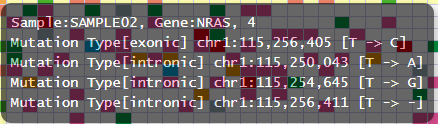
By customizing the configuration file, the information of positions and substitution types can be incorporated.
After customization
Sample,Chr,Start,Ref,Alt,MutationType,Gene
SAMPLE00,chr10,8114472,A,C,intronic,GATA3
SAMPLE00,chr13,28644892,G,-,intronic,FLT3
SAMPLE00,chr13,28664636,-,G,intronic,FLT3
SAMPLE00,chr16,68795521,-,T,UTR3,CDH1
SAMPLE00,chr10,8117068,G,T,exonic,GATA3
SAMPLE00,chr3,178906688,G,A,intronic,PIK3CA
SAMPLE00,chr13,28603715,G,-,intergenic,FLT3
SAMPLE00,chr14,103368263,G,C,intronic,TRAF3
In the example data above, the following four (optional) items are incorporated a part from sample ID, gene name, and mutation type (required items).
- Chromosome (Chr)
- Variant start position (Start)
- Reference base (Ref)
- Alternative base (Alt)
First, add these columns to the [result_format_mutation] section in the configuration file.
[result_format_mutation]
col_opt_chr = Chr
col_opt_start = Start
col_opt_ref = Ref
col_opt_alt = Alt
The column names of optional items can be set as col_opt_{keyword} = {actual column name}.
For a more detailed description on keyword, please refer to About keyword.
Then, modify the [mutation] section in the configuration file.
[mutation]
# before customization
# tooltip_format_checker_partial = Mutation Type[{group}]
# after customization
tooltip_format_checker_partial = Mutation Type[{group}] {chr}:{start:,} [{ref} -> {alt}]
Then, execute paplot.
paplot mutation {unzip_path}/example/mutation_option/data.csv ./tmp mutation_option \
--config_file {unzip_path}/example/mutation_option/paplot.cfg
Here, we describe the procedure to customize the pop-up for each element in the main grid. For customizing other pop-ups, please refer the following:
Six types are set for each display location; however the method of writing is identical.
Correspondence between setting items and display
The following can also be used as a special keyword:
| {#number_id}: | the number of mutations per sample |
|---|---|
| {#number_gene}: | the number of mutations per gene |
| {#number_mutaion}: | |
| the number of mutations (Even if the same sample is detected multiple times with the same gene, it counts as 1.) | |
| {#sum_mutaion}: | Total number of mutations |
| {#item_value}: | Value of one item of stacked graph |
| {#sum_item_value}: | |
| Total value of stacked graph | |
Moreover, for a more detailed description of the procedure to set pop-up information, please refer to User defined format.